Generic residue numbering¶
Sequence-based generic GPCR residue numbering schemes1 exist for class A (Ballesteros-Weinstein, BW2 ) B (Wootten3 ), C (Pin4 ), and F (Wang5 ). In these systems, the first number denotes the helix (1-7) and the second the residue position relative to the most conserved position, which is assigned the number 50. For example, 6.51 denotes a residue in transmembrane helix 6, one position after the most conserved residue (6.50). The reference helix conserved positions differ between the GPCR classes.
Recent GPCR crystal structures have revealed frequent helix bulges and constrictions in several transmembrane helices6 . Structural superimposition makes it clear that these cause a gap that offsets all the following residue numbers when compared to an undistorted helix, i.e. the structurally equivalent residues no longer have the same number (Fig. 1).
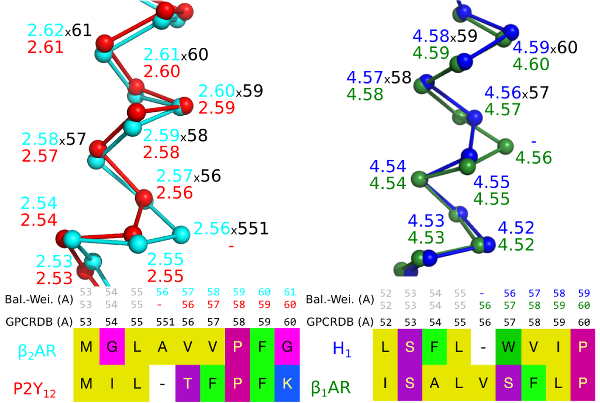
Figure 1. A bulge in helix 2 of the Beta-2 adrenergic receptor (left) and a constriction in helix 4 of the Histamine H1 receptor (right) create offsets in the sequence-based generic numbers when compared to receptors that lack the bulge/constriction.
The GPCRdb numbering scheme1 is the first that is based on crystal structures and corrects for helix bulges and constrictions. GPCRdb numbers are distinguished by a unique separator x and may be used alone, e.g. 5x47, or together with one of the sequence-based schemes, e.g. 5.46x47. A bulge residue is assigned the same number as the preceding residue followed by a 1, e.g. 551 for a bulge following position 55.
GPCRdb offers a suite of tools making it easier to use generic residue numbers:
- Structure-based sequence alignments gapped to account for bulges and constrictions
- Lookup tables with receptor-specific and generic residue numbers
- PDB structure numbering along with visualization tools for PyMOL and Maestro
GPCRdb cross-class alignments contain each of the numbering schemes, which may be distinguished in text by appending the letter of the class, e.g. 2x52ax59b. The Lookup tables tool also provides the alternative class A numbering schemes by Oliveira7 and Baldwin/Schwartz7,8 .
References
- V Isberg et al., 2015, Trends Pharmacol Sci, 36(1), 22–31.
- JA Ballesteros and H Weinstein, 1995, Methods Neurosci, 25, 366–428.
- D Wootten et al., 2013, Proc Natl Acad Sci, 110(13), 5211-5216.
- J-P Pin et al., 2003, Pharmacol Ther, 98(3), 325-354.
- C Wang et al., 2014, Nat Commun, 5, 4355.
- R van der Kant and G Vriend, 2014, Int J Mol Sci, 15(5), 7841-7864.
- L Oliveira et al., 1993, J Comput Aided Mol Des, 7(6), 649–658.
- JM Baldwin, 1993, EMBO J, 12(4), 1693–703.
- TW Schwartz, 1994, Curr Opin Biotechnol, 5(4), 434–44.